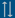| QL-CON-CP1/pFMV2 |
construct-specific |
GT200, GT73, GTSB77, H7-1, J101, J163, MON1445, MON1698, MON87705, MON88302, MON88913, MON89788, RBMT22-082, RBMT22-186, RBMT22-238, RBMT22-262, ZSR500, ZSR502, ZSR503 |
Qualitative conventional PCR method for detection of the junction between the FMV promoter and the cp4-epsps gene (BVL G30.40-11). |
ring trial validation |
national standard |
1 |
| QL-CON-00-008 |
construct-specific |
2904/1 kgs, ASR368, DBN9004, DBN9936, GT200, GT73, GTSB77, H7-1, J101, J163, KWS20-1, MON1445, MON1698, MON71200, MON71700, MON71800, MON80100, MON802, MON809, MON832, MON87411, MON87427, MON87429, MON87705, MON88017, MON88302, MON88913, MON89788, NK603, RBMT22-082, RBMT22-186, RBMT22-238, RBMT22-262, ZSR500, ZSR502, ZSR503 |
Qualitative real-time PCR (TaqMan) method for detection of the junction between the chloroplast transit peptide 2 and the CP4 epsps gene (ISO 21569 AMD1). |
ring trial validation |
ISO standard |
3 |
| QL-CON-35S-F/nptII-R |
construct-specific |
23-18-17, 23-198, 31807, 31808, 5345, 8338, ATBT04-06, ATBT04-27, ATBT04-30, ATBT04-31, ATBT04-36, BT10 potato, BT12 potato, BT16 potato, BT17 potato, BT18 potato, BT23 potato, BT6 potato, BXN10211, CZW-3, EE-1, Event 1, FLAVR SAVR, MON1076, MON1445, MON15985, MON1698, MON531, MON757, MON80100, MON863, MON87460, SP951, SPBT02-7, pap1 GM Gypsophila |
Qualitative real-time PCR (TaqMan) method for detection of the junction between the CaMV 35S promoter and the nptII gene (Reiting, 2010). |
in-house validation |
none |
1 |
| QL-ELE-epsps2 1-5'/epsps2 1-3' |
element-specific |
CS-CP4epsps-RHIRD |
Qualitative real-time PCR (TaqMan) method for detection of the CP4 epsps gene (Mano et al., 2009) |
in-house validation |
none |
3 |
| QL-ELE-npt 1-5'/npt 1-3' |
element-specific |
CS-nptII-ECOLX, V-nptII-ECOLX |
Qualitative real-time PCR (TaqMan) method for detection of the nptII gene (Scholtens et al., 2013). |
in-house validation |
none |
3 |
| QL-ELE-PFMV 1-5'/PFMV 1-3' |
element-specific |
P-34S FMV |
Qualitative real-time PCR (TaqMan) method for detection of P-FMV (Mano et al., 2009) |
in-house validation |
none |
3 |
| QL-ELE-P-FMV-F/P-FMV-R |
element-specific |
P-34S FMV |
Qualitative real-time PCR (TaqMan) method for detection of the P-FMV promoter (Debode et al., 2013) |
in-house validation |
none |
1 |
| QL-ELE-00-001 |
element-specific |
P-Cauliflower mosaic virus |
Qualitative conventional PCR method for detection of Cauliflower mosaic virus 35S promoter (Official Collection of Methods according to § 64 LFGB, L 00.00-31) |
ring trial validation |
national standard |
1 |
| QL-ELE-00-002 |
element-specific |
CS-nptII-ECOLX, V-nptII-ECOLX |
Qualitative conventional PCR method for detection of neomycin phosphotransferase II gene (ISO 21569) |
ring trial validation |
ISO standard |
2 |
| QL-ELE-00-003 |
element-specific |
CS-nptII-ECOLX, V-nptII-ECOLX |
Qualitative conventional PCR method for detection of the neomycin phosphotransferase II gene (nptII) (BVL L 00.00-31, 2001) |
ring trial validation |
national standard |
2 |
| QL-ELE-00-004 |
element-specific |
P-Cauliflower mosaic virus |
Qualitative conventional PCR method for detection of Cauliflower mosaic virus 35S promoter (Lipp et al., 2001) |
ring trial validation |
ISO standard |
2 |
| QL-ELE-00-005 |
element-specific |
P-Cauliflower mosaic virus |
Qualitative conventional PCR method for detection of Cauliflower mosaic virus 35S promoter (ISO/FDIS 21569) |
ring trial validation |
ISO standard |
1 |
| QL-ELE-00-006 |
element-specific |
T-nos-RHIRD |
Qualitative conventional PCR method for detection of nopaline synthase terminator (Official Collection of Methods according to § 64 LFGB, L 00.00-31) |
ring trial validation |
national standard |
2 |
| QL-ELE-00-007 |
element-specific |
T-nos-RHIRD |
Qualitative conventional PCR method for detection of nopaline synthase terminator (EU-Project SMT4-CT96-2072:1998) |
ring trial validation |
national standard |
2 |
| QL-ELE-00-009 |
element-specific |
T-nos-RHIRD |
Qualitative conventional PCR method for detection of nopaline synthase terminator (Lipp et al.,2001) |
ring trial validation |
ISO standard |
2 |
| QL-ELE-00-010 |
element-specific |
E-FMV, P-34S FMV |
Qualitative conventional PCR method for detection of Figwort mosaic virus 34S promoter (Pan et al., 2007) |
ring trial validation |
ISO standard |
2 |
| QL-ELE-00-011 |
element-specific |
T-nos-RHIRD |
Qualitative real-time PCR (TaqMan) method for detection of nopaline synthase terminator (ISO 21569 AMD1). |
ring trial validation |
ISO standard |
3 |
| QL-ELE-00-012 |
element-specific |
E-35S-CaMV, P-Cauliflower mosaic virus, P-scp1-SYNTH |
Qualitative real-time PCR (TaqMan) method for detection of Cauliflower mosaic virus 35S promoter (partim P-35S).
Part of the qualitative duplex real-time PCR (TaqMan) method for detection of Cauliflower mosaic virus 35S promoter and nopaline synthase terminator (ISO 21569 AMD1) |
ring trial validation |
ISO standard |
2 |
| QL-ELE-00-013 |
element-specific |
T-nos-RHIRD |
Qualitative real-time PCR (TaqMan) method for detection of nopaline synthase terminator (partim T-nos).
Part of the qualitative duplex real-time PCR (TaqMan) method for detection of Cauliflower mosaic virus 35S promoter and nopaline synthase terminator (partim T-nos) (ISO 21569 AMD1) |
ring trial validation |
ISO standard |
2 |
| QL-ELE-00-015 |
element-specific |
E-FMV, P-34S FMV |
Qualitative real-time PCR (TaqMan) method for detection of Figwort mosaic virus 34S promoter (ISO/TS 21569-5). |
ring trial validation |
ISO standard |
3 |
| QL-ELE-00-017 |
element-specific |
P-Cauliflower mosaic virus |
Qualitative real-time PCR (SYBRGreen) method for detection of P-35S CaMV (Barbau-Piednoir et al., 2014) |
ring trial validation |
unknown |
2 |
| QL-ELE-00-018 |
element-specific |
T-nos-RHIRD |
Qualitative real-time PCR (SYBRGreen) method for detection of T-nos-RHIRD (Barbau-Piednoir et al., 2014) |
ring trial validation |
unknown |
1 |
| QL-ELE-00-019 |
element-specific |
CS-CP4epsps-RHIRD |
Qualitative real-time PCR (SYBRGreen) method for detection of CS-CP4epsps-RHIRD (Barbau-Piednoir et al., 2014) |
ring trial validation |
unknown |
1 |
| QL-ELE-00-024 |
element-specific |
T-rbcS_E9-PEA |
Qualitative real-time PCR (TaqMan) method for detection of the terminator of the pea ribulose-1,5-bisphosphate carboxylase small subunit (rbcS) E9 gene (EU-RL GMFF GMOMETHODS database as of January 20, 2017). |
ring trial validation |
EU reference method |
3 |
| QT-ELE-NOS ter 2-5' /NOS ter 2-3' |
element-specific |
T-nos-RHIRD |
Quantitative real-time PCR (TaqMan) method for detection of T-nos (Kodama et al., 2009) |
ring trial validation |
none |
3 |
| QT-ELE-P35S 1-5'/P35S 1-3' |
element-specific |
P-Cauliflower mosaic virus |
Quantitative real-time PCR (TaqMan) method for detection of the Cauliflower mosaic virus 35S promoter (Kodama et al., 2009) |
ring trial validation |
national standard |
3 |
| QT-ELE-00-001 |
element-specific |
P-Cauliflower mosaic virus |
Quantitative real-time PCR (TaqMan) method for detection of Cauliflower mosaic virus 35S promoter (Feinberg et al., 2005) |
ring trial validation |
unknown |
1 |
| QT-ELE-00-004 |
element-specific |
E-35S-CaMV, P-Cauliflower mosaic virus, P-scp1-SYNTH |
Quantitative real-time PCR (TaqMan) method for detection of Cauliflower mosaic virus 35S promoter (ISO 21570 Annex B.1) |
ring trial validation |
ISO standard |
3 |
| QT-EVE-GH-003 |
event-specific |
MON1445 |
Quantitative real-time PCR (TaqMan) method for detection of cotton event MON1445 (EU-RL GMFF GMOMETHODS database as of January 13, 2014) |
ring trial validation |
EU reference method |
3 |
| QT-TAX-GH-015 |
taxon-specific |
Gossypium hirsutum |
Quantitative real-time PCR (TaqMan) method for detection of cotton fiber-specific acyl carrier protein (ACP1) gene (EU-RL GMFF GMOMETHODS database as of January 07, 2014).
Estimated number of target copies per haploid genome (Jacchia et al. 2018): 2 |
ring trial validation |
EU reference method |
1 |
| QT-TAX-GH-016 |
taxon-specific |
Gossypium hirsutum |
Quantitative PCR method for detection of putative cotton SAH7 protein gene (123 bp) (Mazzara et al., 2006).
Estimated number of target copies per haploid genome (Jacchia et al. 2018): 2 |
ring trial validation |
EU reference method |
1 |
| QT-TAX-GH-018 |
taxon-specific |
Gossypium hirsutum |
Quantitative real-time PCR (TaqMan) method for detection of cotton alcohol dehydrogenase C (adhC) gene (EU-RL GMFF GMOMETHODS database as of January 17, 2014).
Estimated number of target copies per haploid genome (Jacchia et al. 2018): 1 |
ring trial validation |
EU reference method |
1 |
| QT-TAX-GH-019 |
taxon-specific |
Gossypium hirsutum |
Quantitative real-time PCR (Taqman) method for detection of cotton alcohol dehydrogenase C gene (EU-RL GMFF GMOMETHODS database as of November 1, 2018).
Estimated number of target copies per haploid genome (Jacchia et al. 2018): 1 |
ring trial validation |
EU reference method |
1 |
| QT-TAX-GH-020 |
taxon-specific |
Gossypium hirsutum |
Quantitative real-time PCR (TaqMan) method for detection of cotton alcohol dehydrogenase C (adhC) gene (EU-RL GMFF GMOMETHODS database as of July 2020).
Estimated number of target copies per haploid genome (Jacchia et al. 2018): 1 |
ring trial validation |
EU reference method |
1 |
| QT-TAX-GH-021 |
taxon-specific |
Gossypium hirsutum |
Quantitative real-time PCR (TaqMan) method for detection of putative cotton SAH7 protein gene (115bp) (EU-RL GMFF GMOMETHODS database as January 22, 2014).
Estimated number of target copies per haploid genome (Jacchia et al. 2018): 2 |
ring trial validation |
EU reference method |
1 |